- Components of the reagent kit
| Specifications | 50T | 100T |
| Cat. No. | SN0229 | SN0230 |
| DNA Extraction Columns (set) | 50 (set) | 100 (set) |
| Reagent Buffer B | 20ml | 2×20 ml |
| Reagent Buffer C | 30 ml | 2×30 ml |
| Wash Buffer 1 | 15 ml | 2 × 15 ml |
| Elution Buffer | 20 ml | 20 ml |
| Proteinase K | 1ml | 1ml |
| Lysozyme | 1ml | 1ml |
| RNase A | 1ml | 1ml |
| Instruction Manual | 1 | 1 |
- Storage
This reagent kit should be stored at room temperature (15-25℃) and in dry conditions, with a shelf life of 12 months. The DNA extraction purification columns can be stored for 1 year in a cool and dry environment. Lysozyme, Proteinase K and RNase A contain preservatives, allowing transportation at room temperature, but for long-term storage, they should be kept at -20℃.
- Instructions for Using the Reagent Kit
3.1 This reagent kit is intended for molecular biology research and should not be used for disease diagnosis or treatment.
3.2 Some components in the reagent kit contain irritants. Protective measures such as wearing protective clothing and goggles are recommended.
3.3 During the usage of this reagent kit, a high-speed centrifuge, water bath (metal bath), vortex mixer, anhydrous ethanol, sterile deionized water, and EP tubes need to be prepared by the user.
- Introduction to the Reagent Kit
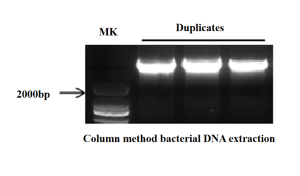
This kit provides a rapid and effective purification method for isolating DNA from various bodily fluids and bacterial culture fluids. It utilizes a silicon-based purification column that selectively adsorbs nucleic acids. With specific buffer solutions, bacterial DNA samples can be extracted within 30 minutes. The entire purification process does not require toxic reagents such as phenol-chloroform. The extracted DNA can be directly used for downstream experiments like PCR, Southern blotting, and others.
- Experimental Principles and Procedures
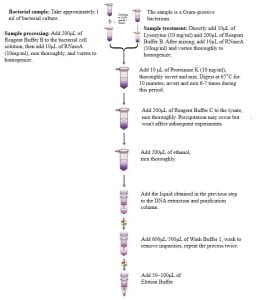
- Extraction Process
Before Starting the Experiment:
- Reagent Buffer B and C may precipitate under low-temperature conditions. We recommend heating at 65℃for 5 minutes. After the precipitate dissolves, it can be used normally.
- Before use, add the specified amount of anhydrous ethanol to Wash Buffer 1as indicated on the bottle label. Mark a check on the label to indicate the addition of anhydrous ethanol.
- Elution Buffer is a0.1x TE solutioncontaining minimal amounts of EDTA. If EDTA might affect subsequent experiments, it is advisable to substitute Elution Buffer with sterile deionized water.
- Sample Handling:
- Take around 1 ml of bacterial culture, centrifuge at 12,000 rpm for 1 minute, aspirate the supernatant as much as possible, add 200μl of Reagent Buffer Bto the bacterial cell solution. Generally, residual RNA has minimal impact on downstream experiments. If RNA interference needs to be eliminated, add 10μl of RNaseA (10mg/ml) to the mixture, incubate at 37°C for 2 minutes with vortexing during the period.
- If the sample being processed contains Gram-positive bacteria, add 10μl of Lysozyme (10 mg/ml)and 200μl of Reagent Buffer B. Generally, residual RNA has minimal impact on downstream experiments. If RNA interference needs to be eliminated, add 10μl of RNaseA (10mg/ml) to the mixture, incubate at 37°C for 15-30 minutes with vortexing during the period.
- Add 10μl of Proteinase K (10 mg/ml), thoroughly invert and mix, digest at 65°C for 2 minutes. During this period, invert and mix the sample solution 6-7 times until the sample solution becomes clear after digestion.
- Add 200μl of Reagent Buffer Cto the lysate and mix. If a white precipitate appears, it can be left to settle; it won’t affect subsequent experiments once the precipitate disappears.
- Add 200μl of ethanol, mix thoroughly. Some precipitation might occur but won’t affect subsequent experiments.
- Transfer the obtained liquid into a DNA extraction purification column, leave at room temperature for 2 minutes, centrifuge at 12,000 rpm for 30 seconds. Discard the collected waste and reinsert the collection tube into the purification column for the next step.
- Add 600μl of Wash Buffer 1, centrifuge at 12,000 rpm for 30 seconds, discard the waste, and reinsert the DNA extraction purification column into the holder.
(Note: Ensure ethanol has been added to Wash Buffer 1.)
- Add 500μl of Wash Buffer 1 to the DNA extraction purification column, centrifuge at 12,000 rpm for 2 minutes, extending the centrifugation time as needed for a drier membrane.
- Place the DNA extraction purification column (holder) into a new centrifuge tube, open, and heat at 65°C for 2 minutes. Extend this step as necessary to evaporate ethanol, preventing ethanol residues from affecting downstream experiments.
- Add 50-100μl of Elution Bufferonto the column membrane, centrifuge at 12,000 rpm for 2 minutes.
(Note: 1. Eluting DNA with 50 μl of Elution Buffer can increase DNA concentration but reduce the total DNA yield. 2. The eluted DNA from the eluate can be reapplied to the DNA extraction purification column, centrifuge again at 12,000 rpm for 2 minutes to enhance DNA yield.)







Reviews
There are no reviews yet.