This study employed the optimized cooking extraction technique to isolate gallus genomic DNA. Analysis via the SmartspecTMplus test revealed an OD260/OD280 ratio of 1.77 ± 0.06, indicating high-quality DNA. PCR amplification confirmed the presence of clear and distinct DNA fragments, affirming the efficacy of this method. In contrast to the Phenol-Chloroform extraction method, the optimized cooking extraction is advantageous due to its absence of chloroform, phenol, and other hazardous substances. Moreover, it stands out for its efficiency in saving time and reducing costs, establishing it as a superior method for genomic DNA extraction.
Since Watson J.D. and Crick F. proposed the double helix model of DNA in the 1950s, molecular biology has experienced unprecedented development in both breadth and depth. Modern molecular biology techniques have permeated various fields such as biology, medicine, genetics, and zoology. The extraction of genomic DNA is a fundamental technique in molecular biology research and a crucial step for subsequent experiments. Currently, there are numerous methods for extracting genomic DNA. However, commercial kits are expensive and not suitable for extracting DNA in larger quantities. Other methods primarily involve organic solvent extraction, which is time-consuming and uneconomical. Therefore, considering the specific requirements of experiments, sample sizes, and economic factors, it is necessary to find a simple, safe, and cost-effective method. Based on reference to several commonly used DNA extraction methods, this study optimized a rapid, simple, efficient, and economical extraction method. This method aims to establish a foundation for research in avian molecular genetics.
1.Materials and Methods
1.1 Materials and Reagents Genomic DNA was extracted from the blood of F2 generation populations of Gushi and Anka chickens. Primers ; Proteinase K, balanced phenol; Taq DNA Polymerase, dNTPs etc.
1.2 Genomic DNA Extraction
1.2.1 Phenol-Chloroform Method: 80 μL of anticoagulant was taken in a 1.5 mL centrifuge tube, added with 500 μL TE buffer, 15 μL ρ (SDS) = 25%, and 15 μL Proteinase K (10 mg/mL), thoroughly mixed, and digested overnight in a 57.8°C water bath. Equal volumes of Tris-saturated phenol were added slowly, shaken slowly for 10 min, centrifuged for 10 min (12,000 rpm); the supernatant was transferred to another centrifuge tube, followed by adding 500 μL Tris-saturated phenol, shaken slowly for 10 min, and centrifuged for 10 min (12,000 rpm). Subsequently, the supernatant was transferred to another centrifuge tube, added with (Vphenol: Vchloroform: Visoamyl alcohol = 24: 23: 1) 600 μL, shaken slowly for 10 min, centrifuged for 10 min (12,000 rpm); the upper layer was taken to another centrifuge tube, added with (Vchloroform: Visoamyl alcohol = 23: 1) 600 μL, shaken slowly for 10 min, centrifuged for 10 min (12,000 rpm). Finally, the supernatant was transferred to another centrifuge tube, added with 2 times the volume of anhydrous ethanol pre-cooled to -20°C to precipitate DNA, shaken slowly for 10 min, centrifuged for 10 min (12,000 rpm), carefully discarded the liquid part; washed with 70% ethanol, shaken slowly for 10 min, centrifuged for 15 min (12,000 rpm); air-dried (inverted); dissolved in 100 μL TE overnight (approximately 24 hours).
1.2.2 Boiling Method: 100 μL of anticoagulant was taken in a 1.5 mL centrifuge tube, added with 1 mL double-distilled water, thoroughly mixed, after 10 min in an ice bath, centrifuged at 13,000 rpm for 1 min, discarded the supernatant, added 100 μL double-distilled water to fully suspend the precipitate, boiled for 5 min, immediately cooled in an ice bath for 5 min, centrifuged at 13,000 rpm for 10 min, and the supernatant was used for PCR detection in another centrifuge tube.
1.2.3 Optimized Boiling Method: Replaced the 100 μL double-distilled water in the boiling method with 100 μL TE.
1.3 Primer Design Using the Primer 5 analysis software, primers were designed for the coding region sequence (second exon) of the chicken PPARα gene (GenBank accession number: NC_006088) based on the genomic sequence and mRNA sequence of the PPARα gene (GenBank accession number: NM_001001464). The primers were synthesized by Shanghai Bioengineering Technology Co., Ltd. Their sequences are: Upstream: 5′-GTTGTTGTTGTGGGTTTAGC-3′; Downstream: 5′-GCAACGTGATTTAGGCTCTT-3′.
1.4 PCR Reaction System and Amplification Conditions
1.4.1 PCR Reaction System: 10x PCR Buffer, 2.5 μL Taq DNA Polymerase (0.5 U/μL) 0.3 μL, dNTPs (10 mmol/mL) 0.5 μL, primers (10 pmol/L) 0.5 μL each, DNA template (100 ng/μL) 1 μL, supplemented with sterile ultrapure water to a final volume of 25 μL.
1.4.2 PCR Reaction Procedure: Pre-denaturation at 95°C for 5 min, denaturation at 94°C for 30 s, annealing at 60°C for 35 s, extension at 72°C for 30 s, 34 cycles, final extension at 72°C for 10 min, and finally stored at 4°C.
2.Results and Analysis
2.1 Nucleic Acid Protein Quantification Results As the quality and yield of DNA extraction directly impact PCR amplification, restriction enzyme digestion, and the success of experiments involving single nucleotide polymorphisms, there is a high requirement for the quality of extracted DNA. The OD260/OD280 value for high-quality DNA typically ranges from 1.7 to 2.0. The results obtained from the nucleic acid protein quantification instrument are shown in Table 1. Both the optimized boiling method and the phenol-chloroform method yielded DNA with OD260/OD280 values within the range of 1.7 to 2.0. However, the DNA extracted using the boiling method had an OD260/OD280 value of only 1.26. While the phenol-chloroform method resulted in the highest DNA concentration, its drawback lies in its time-consuming nature.
Tab. 1 Concentration,purity and blood volume of extracted DNA by three kinds of method
| Extraction method | Concentration/ (ng·uL-1 ) | Purity OD260 /OD280
| Blood volume/ μL |
| Phenol-chloroform extraction | 328. 2 ± 32 . 95 | 1. 89 ± 0 . 11 | 80 |
| Cooking extraction method | 165. 6 ± 15 . 62 | 1. 26 ± 0 . 10 | 100 |
| Optimized cooking extraction method | 204. 7 ± 18 . 13 | 1. 77 ± 0 . 06 | 100 |
2.2 Detection of Genomic DNA Extraction Results
The genomic DNA extracted via the phenol-chloroform method, boiling method, and optimized boiling method was simultaneously subjected to agarose gel electrophoresis, and the results are shown in Figure 1. The results indicate that under the conditions of 10 g/kg agarose gel electrophoresis, after a period of electrophoresis, the genomic DNA obtained from the phenol-chloroform method appeared as a neat, clear, single band without trailing, while no bands were observed for the genomic DNA extracted using the boiling method. This might be attributed to a fraction of the double-stranded DNA undergoing irreversible denaturation under the high-temperature conditions, forming circular coiled DNA. This form of DNA has twice the migration rate in agarose gel electrophoresis compared to normal double-stranded DNA. Consequently, these larger DNA complexes are unable to traverse through the molecular sieve of the agarose gel in the electric field, leading to their retention in the sample wells.
Fig. 1 Comparison of three genomic DNA extraction methods
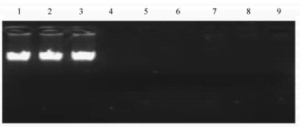
1-3 . phenol -chloroform extraction ; 4 -6 . cooking extraction method ; 7 -9 . optimized cooking extraction method.
2.3 PCR Detection of Genomic DNA Quality
To assess the efficacy of the boiling method, genomic DNA obtained from the three methods was subjected to PCR amplification, and the resulting products were examined by agarose gel electrophoresis (refer to Figure 2). The results indicate that genomic DNA extracted using the three methods exhibited clear, dense bands upon PCR amplification, consistent with expected fragment sizes compared to DNA molecular weight standards. This suggests that the extracted DNA from all three methods meets the requirements for molecular biology experiments.
Fig. 2 Comparison of three genomic DNA extraction methods
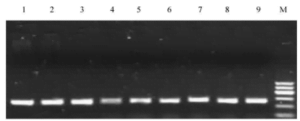
1-3 . Phenol -chloroform extraction ; 4 -6 . cooking extraction method ; 7 – 9 . Optimized cooking extraction method ; M. DNA marker 1,the lightest band is 400 kb.
2.4 PCR Detection of Genomic DNA Quality After One Month of Storage at -20°C
Results indicate that after one month of storage, genomic DNA obtained through the phenol-chloroform method and the optimized boiling method displayed corresponding clear and bright target bands. However, the target bands obtained from genomic DNA extracted using the boiling method appeared fainter, indicating a decrease in the quality of the extracted DNA.
Fig. 3 PCR results of three genomic DNA extraction methods after
being preserved for a month at -20 ℃
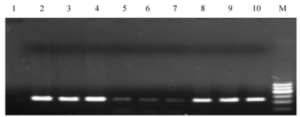
- Negative control ;2-4 . phenol - chloroform extraction ; 5 -7 . cooking extraction method ;8 -10 . optimized cooking extraction method ;M. DNA marker 1,The lightest band is 400 kb.
3.Discussion
- Selection of Extraction Methods: Choosing a method that is quick, simple, efficient, and economical is crucial for genomic DNA extraction. Currently, numerous methods exist, but expensive commercial kits are not suitable for large-scale DNA extraction, and some methods require expensive proteinase K for protein digestion. This experiment explored a concise method for extracting chicken genomic DNA, using osmotic balance principles to break cells, thermodynamic principles to separate chromosomes from fragmented membrane structures, and to isolate genomic DNA from histones, finally obtaining genomic DNA by centrifugation.
- Effects of Freezing on Genomic DNA: To assess the impact of freezing on genomic DNA, PCR amplification was performed on DNA extracted using three methods after one month. Results showed that DNA extracted through the optimized boiling method, like the phenol-chloroform method, remained viable for long-term storage under freezing conditions. However, DNA extracted using the boiling method displayed fainter bands after one month, likely due to a decrease in DNA synthesis purity affecting PCR product quality. While sterile double-distilled water could replace TE for DNA extraction, TE’s EDTA component neutralizes metal ions, inhibiting nucleases and extending storage time. Moreover, TE buffer at a pH of around 8 maintains DNA in a salt ion state, aiding solubility and allowing for increased DNA yield. Thus, for long-term experimental research using genomic DNA, it’s preferable to use TE buffer for storage.
- Comparison of Extraction Methods: Conventional phenol-chloroform extraction yields the best template quality with longer storage times, but it involves complex procedures, high costs, and both phenol and chloroform pose health hazards and environmental risks due to their volatile toxicity. The boiling method saves time and costs but results in higher protein content in the extracted DNA, potentially interfering with DNA polymerase activity for subsequent amplification. On the other hand, the optimized boiling method not only saves time and costs but also yields high-quality DNA with an extended storage period, offering a rapid and straightforward approach for genomic DNA extraction.
