Taq ADN polymérase
Numéro d'article:E001A Spécification:500U/1000U/5000U Storage: Store at -20°C
Présentation du produit:
This product is Taq DNA Polymerase, commonly referred to as Taq enzyme, which is one of the most widely used DNA polymerases. It is expressed in Escherichia coli with the cloned gene of Thermus aquaticus DNA Polymerase and then purified through multiple steps. Le RAP products amplified using this product have an added A base at the 3′ fin, allowing for cloning into T vectors.
Contenu du produit:
| Composants | E001A-01(500U) | E001A-02(1000U) | E001A-03(5000U) |
| Taq DNA Polymerase(5U/µL) | 100µL | 200µL | 1ml |
| dNTP Mixture (10mM chacun) | 100µL | 200µL | 1ml |
| 10× PCR Buffer (Mg2+ plus)
| 1ml | 2× 1mL | 10× 1mL |
Stockage:
Store at -20°C, with a minimum shelf life of 12 mois.
Activity Definition:
Using activated largemouth bass sperm DNA as a template/primer, the intake of 10 nmol of total nucleotides as acid-insoluble material within 30 minutes at 74°C is defined as 1 unit of activity (U).
Contrôle de qualité:
This product has undergone quality testing and is free from endonuclease activity, activité exonucléase, and ribonuclease contamination. Residual host genomic DNA is below 10 copies.
Product Usage:
Amplification of DNA using PCR method; DNA sequence determination.
Instructions d'utilisation:
- Dissolve and mix all required solutions for the PCR reaction and place them on an ice bath or in a cooler. It is recommended to aliquot the PCR reaction mixture to avoid repeated freeze-thaw cycles.
- It is advisable to set up the PCR reaction system on an ice bath or in a cooler, following the recommended reaction system for reference.
- Excessive template DNA can lead to nonspecific PCR products. Recommended template amounts for different types of templates in a 50µL reaction volume are as follows:
Genomic DNA from Animals and Plants: 0.1-1µg;
Genomic DNA from Escherichia coli: 10-100ng;
ADN plasmidique: 0.1-10ng.
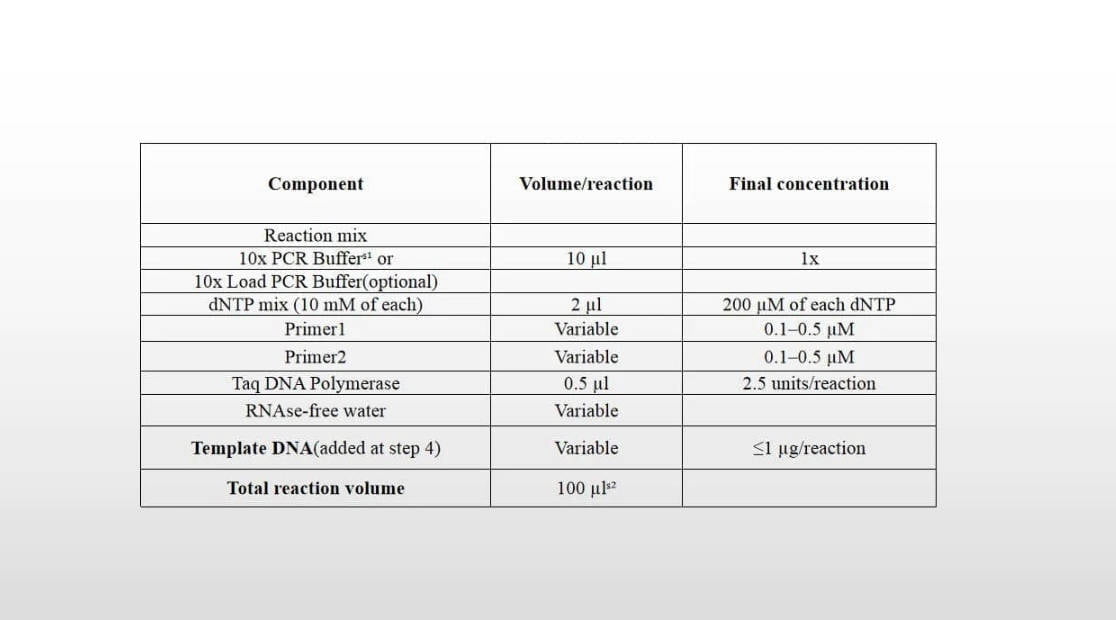
Recommended Reaction System:
| Réactifs | 50µL System Volume | Final Concentration |
| Taq ADN polymérase | 1µL | 0.1U |
| 10× PCR Buffer (Mg2+ plus) | 5µL | 1× |
| dNTP Mixture (10mM chacun) | 1µL | 0.2mM chacun |
| Primer I (10µM) | 1µL | 0.2µM |
| Primer II (10µM) | 1µL | 0.2µM |
| Template DNA | 1µL | — |
| ddH2O | To 50µL | — |
Note: The amounts of each component in the reaction system can be adjusted according to actual requirements.
- Thoroughly pipette and mix the prepared reaction system using a pipette, seal the PCR tubes with caps, label them appropriately, and briefly centrifuge at room temperature.
- Place the prepared PCR tubes into the Appareil PCR, set the reaction conditions, and initiate the PCR reaction.
Reaction Conditions:
| Method/Steps | Time setting | Cycles |
| 95℃ (Pre-denaturation) | 2-5min | 1 |
| 95℃ (Dénaturation) | 30sec | 25-35 Cycles |
| 55℃-60℃ (Recuit) | 30sec | |
| 72℃ (Extension) | 1min | |
| 72℃ (Prolongation finale) | 10min | 1 |
| 4℃ (Hold) | ∞ | — |
Note: Reaction conditions can be adjusted and optimized according to actual requirements.
Précautions:
- Extension time can be adjusted based on factors such as the length and GC content of the PCR product. The extension time per kb of the product is closely related to the complexity of the template: simple templates require 20 secondes, typical templates require 30 secondes, and complex templates require 1 minute.
- The setup of the PCR reaction should be tailored to different conditions such as template, primers, length of PCR product, and GC content. The final concentration of primers typically falls within the range of 0.1-1.0μM. DNA template concentration can be adjusted accordingly. For complex templates or high GC content, it’s recommended to prolong pre-denaturation/denaturation or extension times and increase denaturation/annealing temperatures.
- Use designated areas and pipettes before and after amplification, wear gloves, and change them frequently. After completing the PCR reaction, do not open the reaction tubes immediately. Allow them to cool sufficiently at 4°C or -20°C before opening to minimize the risk of PCR product contamination to the laboratory environment.








Commentaires
Il n'y a pas encore de critiques.