Overview of Soil Microorganisms:
- Microbial Diversity: Soil samples contain a vast number of microorganisms, most of which cannot be directly cultivated for research.
- Importance of DNA Extraction: Extracting DNA is a crucial method for studying these microorganisms in soil samples.
DNA Extraction Methods:
- Direct Method:
- Process: Involves placing soil samples directly into a lysis solution and using mechanical or chemical methods to break cell walls, thereby releasing microbial DNA into the solution.
- Techniques: Includes wall-breaking methods for effective lysis; for example, Zhou’s method.
- Advantages: Maximizes the likelihood of obtaining the entire genomic content (metagenome) of soil microorganisms.
- Indirect Method:
- Process: Begins with suspending soil in a buffer solution (e.g., Buffer PBS) to separate microorganisms from soil particles before DNA extraction.
- Advantages: Reduces the impact of inhibitors like humic acids and heavy metal salts on the DNA extraction process.
- Disadvantages: May result in the loss of many microorganisms during separation, and typically does not recover the complete metagenome.
Challenges and Considerations:
- Direct Method Challenges: Although it increases the chances of obtaining complete genomes, this method can be compromised by the presence of inhibitors such as humic substances and heavy metals in the soil.
- Indirect Method Limitations: Fewer researchers use this method due to its potential to lose microbial diversity and completeness of the genomic extraction.
- Humic acid pollution.The soil, especially in forests and grasslands, is rich in humic acids. Humic acid is a series of organic molecules, some of which are very similar to nucleic acid molecules and difficult to remove during purification. Trace amounts of humic acid pollution can lead to downstream applications such as PCR and enzyme digestion failure.
- Lysis method.Soil samples contain various microorganisms, such as bacteria and fungi. Gram positive bacteria and fungi both contain very thick bacterial walls, and effectively breaking down the cell walls of these microorganisms is crucial for extracting high-yield metagenomic DNA. Due to the complexity of soil samples, it is not feasible to use enzymatic methods (such as lysozyme, wall breaking enzyme, snail enzyme) or liquid nitrogen grinding, as the soil contains various metalions or inhibitory factors that inactive the digestive enzymes, or the presence of sand particles in the soil makes liquid nitrogen grinding difficult.
- The DNA yield is difficult to control.Soil samples would have significant changes in the number and variety of microorganisms due to fertility, inferiority, high moisture content, dryness, or depth of sampling. In a small range of soil samples, the DNA content often varies by thousands of times. In addition, certain chemical components in soil, such as heavy metal salts and clay substances, can cause a decrease in DNA yield.
Features of Magen’s HiPure Soil DNA Kits:
-
Optimization:
- Currently, considered the most optimized kit for soil DNA extraction, providing efficient and reliable results.
-
Extraction Methods:
- Utilizes a combination of glass bead grinding and thermal shock chemical wall-breaking methods.
- Compatible with point vortex instruments, eliminating the need for specialized bead grinding equipment.
- Suitable for a wide range of laboratory settings.
-
Humic Acid Removal:
- Incorporates an Absorber Solution, a proprietary humic acid adsorbent developed by Magen.
- Efficiently removes various humic acid pollutants from soil samples, enhancing DNA purity.
-
Soluble Inhibitor Removal:
- Employs an alcohol-free silica gel column purification method to remove soluble metal salts and other inhibitory factors from soil samples.
-
Versatile Application:
- Successfully extracts DNA from various soil types, including forest soil, mangrove soil, grasslands, farmland, seabed mud, sludge, mineral area soil, and more.
- Supports rapid and reliable isolation of high-quality genomic DNA.
-
Processing Capacity and Time:
- Capable of processing up to 500 mg of soil samples within 60 minutes, facilitating high-throughput workflows.
-
PCR Compatibility:
- Purified DNA is suitable for PCR, restriction digestion, and next-generation sequencing applications.
-
Efficiency and Sustainability:
- Utilizes the reversible nucleic acid binding properties of the HiPure matrix combined with spin column technology to eliminate PCR-inhibiting compounds like humic acid.
- Reduces plastic waste and hands-on time by eliminating the need for organic extractions, enabling parallel processing of multiple samples.
Specifications
| Features | Specifications |
| Main Functions | Isolation DNA from 200-500mg soil sample |
| Applications | PCR, southern blot and enzyme digestion, etc. |
| Purification method | Mini spin column |
| Purification technology | Silica technology |
| Process method | Manual (centrifugation or vacuum) |
| Sample type | Soil |
| Sample amount | 200-500mg |
| Elution volume | ≥30μl |
| Time per run | ≤60 minutes |
| Liquid carrying volume per column | 800μl |
| Binding yield of column | 100μg |
Soil DNA Extraction Process:
- Homogenization and Lysis:
- The soil sample is homogenized to ensure uniformity.
- It is then treated in a specially formulated buffer containing detergent to effectively lyse bacteria, yeast, and fungal samples.
- Contaminant Removal:
- Our proprietary Absorber Solution efficiently removes humic acid, proteins, polysaccharides, and other contaminants from the lysed sample.
- Binding and Washing:
- The lysed sample undergoes adjustment of binding conditions and is applied to a DNA Mini Column.
- Two rapid wash steps effectively remove trace contaminants, ensuring purity.
- Elution of Purified DNA:
- Pure DNA is eluted from the column in a low ionic strength buffer.
- The purified DNA obtained is of high quality and can be directly used in downstream applications without the need for further purification.
Advantages
- Fast – several samples can be extracted in 40 minutes (after digestion)
- High purity – purified DNA can be directly used in various downstream applications
- Good repeatability – silica technology can obtain ideal results every time
- High recovery – DNA can be recovered at the level of PG
Kit Contents
| Contents | D314202 | D314203 |
| Purification Times | 50 Preps | 250 Preps |
| Hipure DNA Mini Columns II | 50 | 250 |
| 2ml Collection Tubes | 50 | 250 |
| 2ml Bead Tubes | 50 | 250 |
| Buffer SOL | 60 ml | 250 ml |
| Buffer SDS | 5 ml | 20 ml |
| Buffer PS | 10 ml | 50 ml |
| Absorber Solution | 10 ml | 50 ml |
| Buffer GWP | 40 ml | 220 ml |
| Buffer DW1 | 30 ml | 150 ml |
| Buffer GW2* | 20 ml | 2 x 50 ml |
| Buffer AE | 15 ml | 30 ml |
Storage Instructions:
- Absorber Solution:
- Upon arrival, store at 2-8°C.
- Short-term storage (up to 24 weeks) at room temperature (15-25°C) does not affect performance.
- Remaining Kit Components:
- Store dry components at room temperature (15-25°C).
- Stable for at least 18 months under these conditions.
Experiment Data
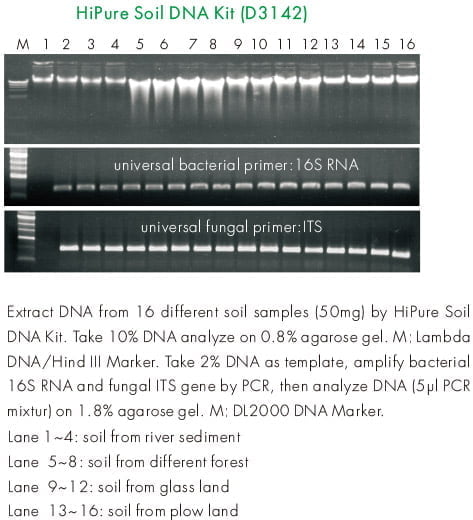



Reviews
There are no reviews yet.