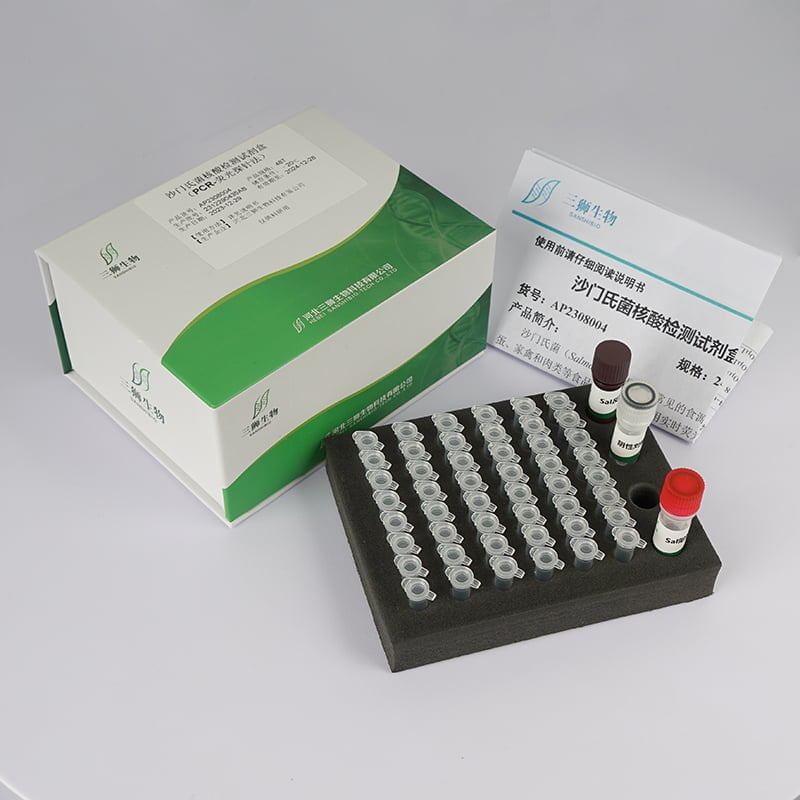
Item No:AP2308004 Specifiation:24T/48T Storage:-20℃
Product introduction:
Salmonella is a common foodborne pathogenic bacterium that can lead to salmonellosis, with eggs, poultry, and meat being common sources of transmission. This test kit utilizes real-time fluorescent quantitative PCR technology to selectively amplify specific DNA segments of Salmonella in samples such as water, feces, and suspected contaminated food. The presence of Salmonella in the sample can be determined by the Ct value, or the contamination level of Salmonella in the sample can be determined through a standard curve.
Product contents:
| Reagent components | AP2308004-01(24T) | AP2308004-02(48T) |
| Sal Premix Solution | 11.5µL/well× 8wells/strip× 3strips | 11.5µL/well× 8wells/strip× 6strips |
| Sal Reaction Solution | 290μL | 575μL |
| Sal Positive control | 100μL | 100μL |
| Negative control | 100μL | 100μL |
Storage conditions:
Store at -20℃, the shelf life is at least 12 months.
Experimental operation:
- Sample Preparation (Sample Preparation Area)
1.1 Sample Requirements:
– Oral and cloacal swabs: Take a sterile cotton swab, insert it into the bird’s throat or cloaca, rotate three times, place it in a centrifuge tube, cut the exposed part, tightly close the cap, and label it. Samples that can be tested promptly should be stored at 2-8°C within 24 hours, and samples that cannot be tested promptly should be stored at -20°C.
– Whole blood: Use EDTA as an anticoagulant, avoid heparin anticoagulant, use fresh whole blood, or store at 2-8°C for no more than 7 days, or store at -20°C for no more than 3 months, avoiding repeated freeze-thaw cycles.
– Tissue: Take fresh muscle or organ tissue not exceeding 100mg, use 200-500μL sterile water to prepare tissue homogenate, and proceed with subsequent sample preparation.
1.2 Sample Preparation:
According to the “GB 4789.4-2016 National Food Safety Standard – Food Microbiological Examination – Salmonella Test” section 5.1 or other applicable standards, process the sample by pre-enrichment, and prepare the bacterial suspension for later use. Weigh 25g (mL) of the sample and place it in a sterile homogenization cup containing 225mL of Buffered Peptone Water (BPW). Homogenize at 8000rpm/min to 10000rpm/min for 1min to 2min or place it in a sterile homogenization bag containing 225mL of BPW and homogenize with a stomacher for 1min to 2min. If the sample is liquid, homogenization is not necessary; shake it well. If pH measurement is required, adjust the pH to 6.8±0.2 using 1mol/mL sterile NaOH or HCl.Perform aseptic operations to transfer the sample to a 500mL conical flask. If using a homogenization bag, direct cultivation can be performed. Incubate at 36°C±1°C for 8h to 18h. If the sample is a frozen product, thaw at temperatures below 45°C for no more than 15min or at 2°C to 5°C for no more than 18h.Refer to the Sanshi Biological “Total DNA/RNA Extraction Kit Manual” (Catalog: DP201) or other nucleic acid extraction kits/methods that meet relevant requirements for nucleic acid extraction from processed samples. Place the extracted nucleic acid samples in an icebox and perform the detection as soon as possible. Store at 4°C for no more than 7 days or at -20°C for no more than 6 months.
- Reaction System Preparation (Reaction Area)
2.1 Take out each component of the kit, completely thaw at room temperature, centrifuge for 10 seconds, and centrifuge the liquid inside the tube wall and tube cap to the bottom of the tube. Prepare the reaction solution according to the sample quantity (n+2, where n is the number of samples, it is recommended to set up 1 positive control and 1 negative control for each experiment). Specific preparation method: Take (n+2) reaction tubes containing Sal premix solution, add 11.5µL of PiHV reaction solution to each tube, use a pipette to thoroughly mix, 23µL per well, and store in a cooler.
2.2 Then sequentially add 2μL of negative control, extracted sample nucleic acid, and Sal positive control to the reaction solution. Close the tube cap, make a record, and the total volume of each reaction is 25μL. Mix thoroughly, centrifuge for 10 seconds, and perform amplification experiments on the PCR instrument.
- PCR Amplification (Amplification and Product Analysis Area)
Pre-denaturation at 95°C for 2 minutes; Denaturation at 95°C for 10 seconds, annealing and extension at 60°C for 35 seconds, 40 cycles. Collect fluorescence signals at 60°C. Choose the FAM fluorescence channel (use real-time fluorescence quantitative PCR instruments such as ABI series, if necessary, contact the manufacturer or add ROX correction dye; otherwise, follow normal procedures).
- Result Interpretation
4.1 Conditions for Experiment Validity:
Positive control: Ct value in the FAM channel < 28 and the appearance of an “S”-shaped amplification curve. Negative control: No Ct value or Ct value ≥ 40 and no “S”-shaped amplification curve. The experiment results are valid under these conditions; otherwise, repeat the experiment. If repeated testing is still invalid, please contact the manufacturer’s technical personnel.
4.2 Interpretation of Sample Results:
Ct value ≤ 35 with an “S”-shaped amplification curve is considered positive for Salmonella nucleic acid. Ct value between 35 and 40 is considered suspicious, and retesting is recommended. This may be due to substandard nucleic acid extraction or the presence of strong inhibitory substances (such as residual disinfectants, anticoagulants, etc.). It is suggested to reprocess the sample for nucleic acid extraction and amplification. First, exclude “false-positive” results caused by aerosol contamination, then re-extract and test the nucleic acid. If, after excluding aerosol contamination, the FAM channel retest shows a Ct value < 35 with a clear amplification curve, it is considered positive; otherwise, it is considered negative.
No Ct value or Ct value ≥ 40 and no “S”-shaped amplification curve is considered negative for Salmonella nucleic acid.
Precautions:
- To prevent contamination, the experiment should be strictly partitioned, and physical isolation between partitions is preferred to avoid cross-contamination due to human factors. Wear lab coats and latex gloves during the experiment, use tools independently in different areas, change gloves and lab coats when needed. After PCR, do not open the lid immediately; open it after sufficient cooling to minimize aerosol contamination.
- Thaw the reagents completely before use, but avoid repeated freeze-thaw cycles. The PCR reaction tubes provided in the kit are 0.2mL; if replacement is needed, transfer after complete thawing. Strictly follow the instructions for reagent preparation and sample addition. Workstations, centrifuges, pipettes, and other instruments should be disinfected regularly with chlorine-containing disinfectants, alcohol, nucleic acid decontamination agents, or UV light.
- A negative result does not necessarily mean the host is not infected with the virus. Poor sample quality, low virus load, or the presence of strong inhibitory substances (such as alcohol, disinfectants, anticoagulants, etc.) may result in unsuccessful nucleic acid extraction (detection) and a “negative” result. Follow the result interpretation criteria, and when there are suspicious results, first exclude aerosol contamination. If there are other questions, contact the manufacturer’s technical personnel.
- This product is for one-time use only. The components in the reagent are sensitive to natural light; avoid light exposure during packaging and storage. After packaging, do not repeatedly freeze-thaw. For scientific research use only; not for clinical diagnosis or other purposes.



Reviews
There are no reviews yet.