2× Taq PCR Master Mix(without dye)
Item No: E003 Specifiation:1mL Storage: Store at -20°C
Product introduction:
The 2x Taq PCR Master Mix is a premixed solution at twice the concentration of DNA Polymerase, PCR Buffer, and dNTP Mix. When using this product, simply add the template and primers to the premixed solution, and complete the volume with ddH2O for the PCR reaction. This simplifies the operational steps and reduces the risk of contamination during PCR procedures. This product offers high sensitivity, strong specificity, and a wide linear range, allowing for more accurate quantification of target genes. The PCR products amplified using this product have an added A base at the 3′ end, facilitating cloning into T vectors.
Product contents:
| Components | E003 |
| 2× Taq PCR Master Mix(without dye) | 1mL |
Storage:
Store at -20°C, with a minimum shelf life of 12 months.
Quality Control:
This product has undergone quality testing and is free from endonuclease activity, exonuclease activity, and ribonuclease contamination. Residual host genomic DNA is below 10 copies.
Product Usage:
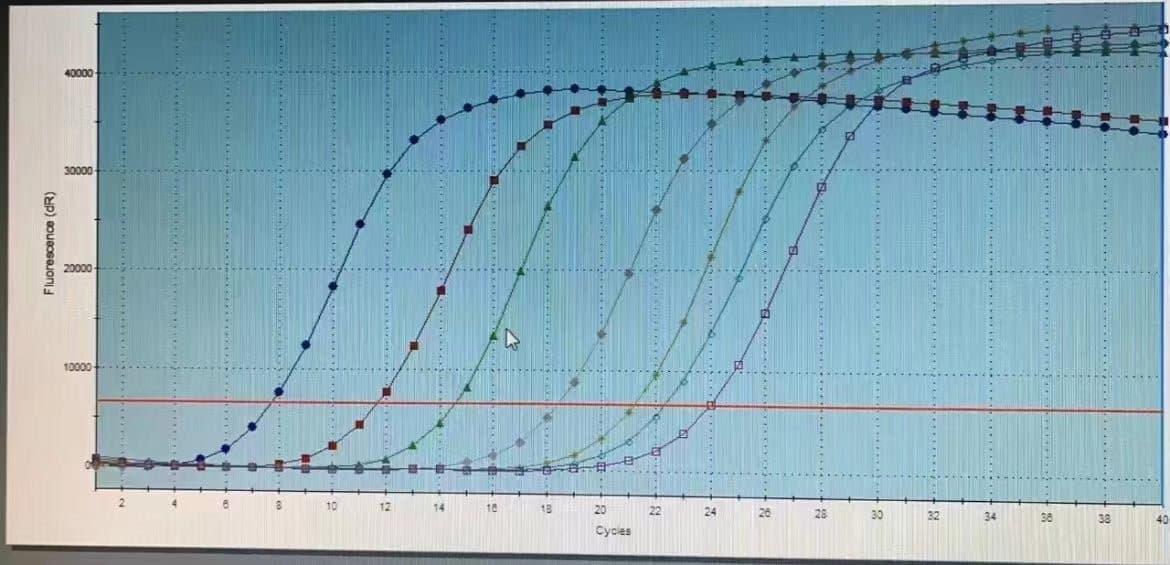
Amplification of DNA using PCR method; DNA sequence determination.
Usage Instructions:
- Allow all required PCR reaction solutions to equilibrate to room temperature until fully dissolved. Thoroughly mix the solutions and set up the PCR reaction system on ice.
- Setting up the PCR reaction system on an ice bath or in a cooler is recommended, following the recommended reaction system for reference.
- Thoroughly pipette and mix the prepared reaction system using a pipette, seal the PCR tubes with caps, label them appropriately, and briefly centrifuge at room temperature.
- Place the prepared PCR tubes into the PCR machine, set the reaction conditions, and initiate the PCR reaction.
Recommended Reaction System:
| Reagents | 25µL System Volume | Final Concentration |
| 2× Taq PCR Master Mix(without dye) | 12.5μL | 1× |
| Primer I (10µM) | 0.5-2.5μL | 0.2-1.0μM |
| Primer II (10µM) | 0.5-2.5μL | 0.2-1.0μM |
| Template DNA | 1μL as required | — |
| ddH2O | Up to 25μL | — |
Reaction Conditions:
Under typical circumstances, a two-step method can be used for the reaction. If the two-step amplification is not optimal, a three-step method can be employed to set up the PCR reaction program. Considering the uncertainty in target gene abundance within the cDNA template, it is recommended to use 35-38 cycles for the initial amplification and adjust the number of cycles based on the amplification results.
| Method/Steps | Time setting | Cycles |
| 95℃ (Pre-denaturation) | 2-5min | 1 |
| 95℃ (Denaturation) | 10sec | 28-40Cycles (Plasmid or Genomic Template:28-35cycles; cDNA Template: 30-40cycles) |
| 55℃-60℃ (Annealing) | 30sec | |
| 72℃(Extension) | 1min / 1-3kb | |
| 72℃ (Final Extension) | 5-10min | 1 |
| 4℃/16℃(Hold) | ∞ | — |
Note: Reaction conditions can be adjusted and optimized according to actual requirements.
Precautions:
- Extension time can be adjusted based on factors such as the length and GC content of the PCR product. The extension time per kb of the product is closely related to the complexity of the template: simple templates require 20 seconds, typical templates require 30 seconds, and complex templates require 1 minute.
- The setup of the PCR reaction should be tailored to different conditions such as template, primers, length of PCR product, and GC content. The final concentration of primers typically falls within the range of 0.2-1.0μM. DNA template concentration can be adjusted accordingly. For complex templates or high GC content, it’s recommended to prolong pre-denaturation/denaturation or extension times and increase denaturation/annealing temperatures.
- Use designated areas and pipettes before and after amplification, wear gloves, and change them frequently. After completing the PCR reaction, do not open the reaction tubes immediately. Allow them to cool sufficiently at 4°C or -20°C before opening to minimize the risk of PCR product contamination to the laboratory environment.
*For OEM, please contact to our salesperson







Reviews
There are no reviews yet.