About the research
Currently used internal reference genes in qRT-PCR include various genes like ubiquitin-conjugating enzyme (UBC), ribosomal RNA (rRNA), elongation factor 1-alpha (eEF1-α), ribosomal proteins (RPL), arginine kinase (AK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin, and tubulin. Evaluating their stability is crucial because different species and conditions show variations in the stability of these reference genes.
The Callosobruchus genus includes pests like the Cowpea weevil, which damages seeds of leguminous crops such as mung beans and red beans. These weevils lay eggs on tender bean pods in fields, and when the larvae hatch, they tunnel into the beans. Eggs and larvae can also enter storage after harvesting, causing a second wave of infestation, which can be more severe if not controlled promptly. These pests are adept at hiding, have a wide distribution, and possess a strong ability to cause repeated infestations, significantly impacting the yield and quality of leguminous crops like mung beans and red beans. Although there are studies on the gene expression of these weevils, the exploration of internal reference genes remains incomplete. This study identifies eight candidate internal reference genes based on previous laboratory research: α-tubulin, β-tubulin, β-actin, elongation factor (eEF1-α), arginine kinase (AK), heat shock protein 70 (Hsc70), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and ribosomal protein 40 (RPL40). These genes were assessed for their expression levels using qRT-PCR. Through GeNorm, NormFinder, BestKeeper, and the online tool RefFinder, along with ΔCt values, the study explores the expression stability of these candidate internal reference genes in different developmental stages of the Cowpea weevil and in various tissues of adult weevils. This research aims to provide reliable internal reference genes for future studies on gene expression and function in the Cowpea weevil.
About the test
Total RNA was extracted from SENO animal tissues using a kit.
Different tissues from Cowpea weevils at various developmental stages and adult weevil tissues were used to synthesize cDNA using the HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper).
This process was repeated three times following the kit instructions. The concentration and quality of the extracted total RNA were checked using a spectrophotometer and agarose gel electrophoresis.
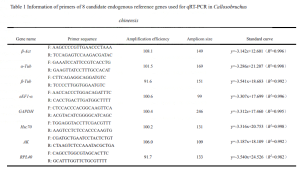
Eight protein-coding genes (β-Act, β-Tub, α-Tub, AK, GAPDH, RPL40, Hsc70, eEF1-α) were chosen as candidate genes.
Quantitative primers were designed using Beacon Designer 7 software and synthesized by a biotech company.
The cDNA templates were diluted in a 1:10 gradient to create five concentrations for qRT-PCR reactions, constructing standard curves, and calculating primer amplification efficiency E.
The qRT-PCR results were used to plot standard curves for the primers, and the amplification efficiency E was calculated using the slope of the standard curve (E = (10^-1/slope^-1) × 100).
The qRT-PCR reaction mixture (20 μL) comprised ChamQ Universal SYBR qPCR Master Mix (10 μL), ddH2O (8.2 μL), PCR Forward Primer (0.4 μL, 10 μmol/L), PCR Reverse Primer (0.4 μL, 10 μmol/L), and cDNA (1 μL). The reaction conditions were: initial denaturation at 95℃ for 30 s; 40 cycles of denaturation at 95℃ for 10 s, annealing/extension at 60℃ for 30 s; and a melting curve from 60 to 95℃ for 30 s. Amplification and melting curves for qRT-PCR were confirmed. Each sample underwent three technical replicates.
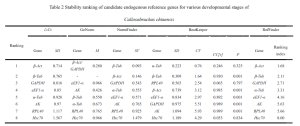
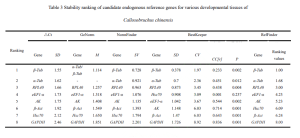
The data from qRT-PCR Ct values were analyzed using △Ct, GeNorm (Vandesompele et al., 2002), BestKeeper (Pfaffl et al., 2004), and NormFinder (Andersen et al., 2004). △Ct and BestKeeper used raw Ct values, while GeNorm and NormFinder transformed the data into 2^-△Ct. △Ct calculated the standard deviation (SD) of Ct value differences between each candidate reference gene pair for stability ranking—smaller SD indicating more stable genes. GeNorm ranked candidate reference gene stability using stability value (M) calculation. When M < 1.5, the gene was considered suitable for reference gene selection, with lower M values indicating higher stability. Moreover, GeNorm determined the optimal number of reference genes using the pairwise variation value V. If the Vn/n+1 value was below the threshold of 0.15, n was deemed the optimal number of reference genes, without the need for adding the n+1th reference gene for correction. BestKeeper software evaluated the stability of candidate reference genes based on Ct value standard deviation (SD), coefficient of variance (CV), correlation coefficient (CC[r]), and significance level (P). Genes with SD < 1 were considered stable, with smaller SD and CV values indicating higher stability. A higher CC[r] value, under P < 0.05, signified a greater correlation with other genes, making it a good candidate as a control gene. Finally, RefFinder, an online tool, comprehensively analyzed reference gene stability, providing a reliable composite ranking index—lower values indicated more stable internal reference genes.
The melting curve analysis from qRT-PCR confirmed that the eight candidate internal reference genes (β-Act, β-Tub, α-Tub, AK, GAPDH, RPL40, Hsc70, eEF1-α) had no non-specific amplification, demonstrating good specificity for subsequent experiments (Figure 1). The results from standard curves for the eight candidate internal reference genes showed a good linear relationship within the dilution gradient, meeting the amplification conditions for qRT-PCR.
Conclusion
Currently, gene expression studies mainly rely on qRT-PCR technology, commonly using reference genes to correct errors introduced during sample detection. Earlier research generally assumed that housekeeping genes essential for maintaining cellular activities exhibit stable expression patterns under different conditions, being regarded as universal reference genes.
However, with the advancement of high-throughput sequencing, more studies suggest that housekeeping genes are not always stable. Optimal reference genes often vary significantly among different species under different conditions.
When studying the expression of target genes, using previously published reference genes without validation can lead to biased results. Hence, it’s essential to reselect reference genes.
In the case of the Cowpea weevil, it was found that the pairwise variation value Vn/n+1 in the GeNorm software’s pairwise difference analysis was consistently less than the default value of 0.15 across different developmental stages and various tissues of adult weevils. Therefore, there is no need to include a third reference gene for data correction; the most suitable number of reference genes is two. Comprehensive assessment through △Ct, GeNorm, NormFinder, BestKeeper, and RefFinder online tool suggests using β-Act and β-Tub as internal reference genes for gene expression studies across different developmental stages of the Cowpea weevil. Similarly, for various tissues in adult weevils, β-Tub and α-Tub were recommended as internal reference genes.
RPL40 was evaluated as relatively suitable for use as one of the internal reference genes in different tissues of adult Cowpea weevils. However, it appeared to be one of the least stable genes among different developmental stages of the weevils.
On the other hand, GAPDH showed the least stability in expression among different tissues of adult Cowpea weevils and was also unsuitable as a reference gene for gene expression studies in different developmental stages of the weevils. Similar results were observed in other Coleopteran insects such as Colaphellus bowringi and Sympiezomias velatus (Tan et al., 2015; Li et al., 2018).
This study provides accurate and reliable internal reference genes for accurately analyzing gene expression levels in different developmental stages and various tissues of the Cowpea weevil using qRT-PCR. However, whether these findings apply to other experimental conditions requires further investigation.
The expression of internal reference genes in the Cowpea weevil varies significantly under different experimental conditions. This study utilized five methods to evaluate the expression stability of eight candidate internal reference genes in different developmental stages and various tissues of the Cowpea weevil. The results suggest that GeNorm software calculated V2/3 gene pairwise difference values under different conditions, all of which were below the threshold of 0.15. Therefore, the optimal number of reference genes for studying relative gene expression is two. Specifically, for different developmental stages of the Cowpea weevil, it is recommended to use β-Act and β-Tub as internal reference genes. Meanwhile, for various tissues in adult weevils, β-Tub and α-Tub are recommended as internal reference genes.
