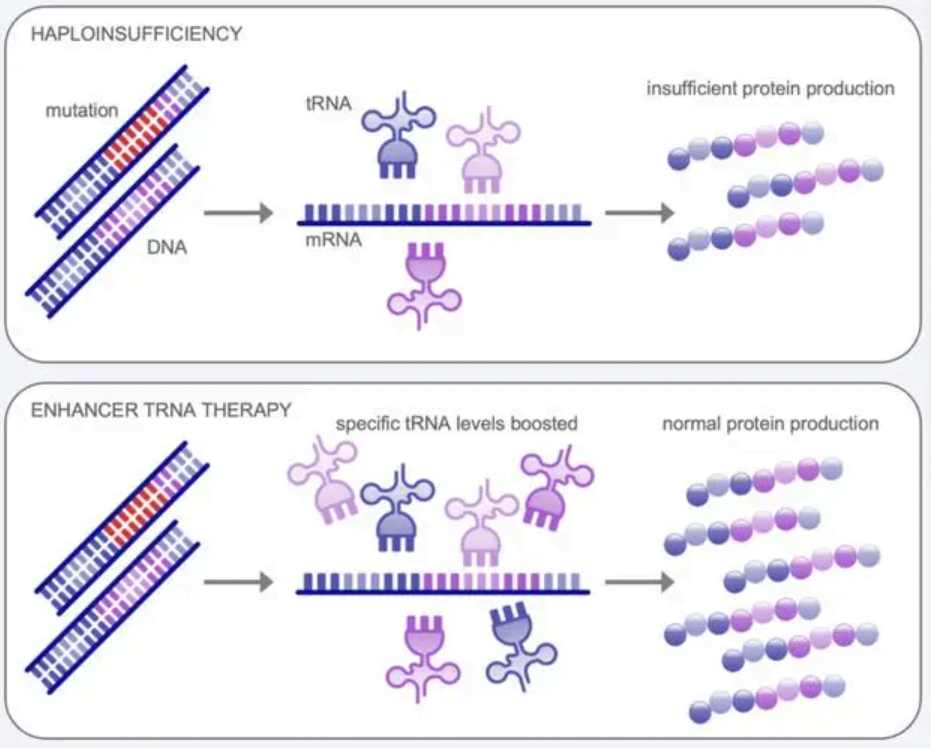
What is tRNA?
Transfer RNA, commonly known as tRNA, plays a vital role in protein synthesis by bridging codons found in messenger RNA (เอ็มอาร์เอ็นเอ) with their corresponding amino acids. Each tRNA molecule contains an anticodon loop that recognizes specific codons through base pairing.
This allows tRNA to deliver the correct amino acid specified by the mRNA codon to the ribosome for protein assembly. There are over 500 different tRNA genes found in the human genome that encode tRNAs carrying all 20 standard amino acids. This ensures there are enough tRNAs available to facilitate efficient translation of the genetic code.
How does tRNA recognize mRNA codons?
The key to tRNA’s function lies in its anticodon, a sequence of three nucleotides that form hydrogen bonds with a complementary mRNA codon. This base pairing follows Watson-Crick complementarity rules, where adenine (ก) pairs with uracil (ยู) and cytosine (ค) pairs with guanine (G).
A given tRNA carries only one type of amino acid, determined by its specific anticodon sequence. ตัวอย่างเช่น, a tRNA with the anticodon UAC would recognize the codon AUG and attach the amino acid methionine. This precise base pairing allows tRNAs to faithfully translate codons and assemble the correct protein sequence.
What is wobble pairing and how does it expand the genetic code?
Some tRNAs exhibit a phenomenon known as wobble pairing, where the third position of the anticodon can form base pairs with more than one nucleotide in the codon. For instance, a guanine (G) in the anticodon third position can pair with either cytosine (ค) or uracil (ยู) in the mRNA codon.
This increased flexibility at the third anticodon position allows a single tRNA to recognize multiple codons specifying the same amino acid. Through wobble pairing, a single tRNA can bind to two or more codons. This enhances translational efficiency by economizing on the number of distinct tRNA molecules needed to translate the genetic code.
How do amino acids get attached to tRNAs?
Aminoacyl-tRNA synthetases catalyze the attachment of specific amino acids to cognate tRNA molecules. Each synthetase recognizes only one tRNA and its corresponding amino acid. The reaction is fueled by hydrolyzing ATP to AMP and pyrophosphate. Each synthetase possesses distinctive structural elements and binding pockets tailored to precisely discriminate between its target tRNA-amino acid pair and all others.
Occasionally mistakes occur during editing of incorrectly attached amino acids. Proofreading steps like hydrolysis of mischarged tRNAs protect against errors and ensure high-fidelity translation of the genetic code.
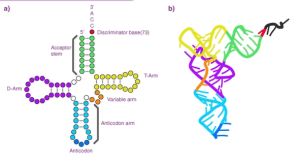
What is the structure of a tRNA molecule?
The primary sequence of a tRNA folds into a distinctive L-shaped three-dimensional structure stabilized by intra-molecular base-pairing. One arm features the anticodon loop while the other end anchors the attached amino acid. Different tRNA transcripts adopt subtly varied structures enabling aminoacyl-tRNA synthetases to distinguish them with exquisite selectivity.
This robust yet adaptable architecture perfectly equips tRNA to undergo amino acid charging and delivery to the ribosome. Molecular modeling has provided insights into structural rearrangements that may accompany tRNA’s functional transitions during protein synthesis.
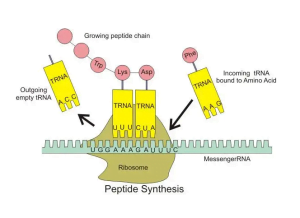
How do tRNAs interact with the ribosome?
After aminoacylation, tRNAs interact with mRNA and the ribosome to facilitate protein assembly. The ribosome contains three binding sites for tRNAs – the A, P and E sites. Charged tRNAs enter the A site and pair with cognate codons. Catalyzed by elongation factors, peptide bond formation links the incoming amino acid to the growing polypeptide chain followed by translocation of tRNAs to the P and E sites.
Empty tRNAs exit the ribosome from the E site to be recharged for subsequent rounds of translation. Precisely coordinated movements of mRNA, tRNAs and ribosomal subunits drive the protein production factory.
How are tRNAs transcribed?
tRNAs are transcribed from genes in the nucleus by RNA polymerase III. tRNA genes contain an internal promoter region recognized by RNA polymerase III and other transcription factors. The initial transcript is a long precursor tRNA containing extra sequences that need to be processed.
This precursor undergoes 5′ capping, intron splicing, and 3′ trailer addition. Enzymes make precise cuts to trim off flanking sequences and ligate exons, leaving only the canonical cloverleaf secondary structure.
What role do modification enzymes play?
After transcription and processing, tRNAs undergo extensive post-transcriptional modification catalyzed by various modification enzymes. เกิน 100 different modifications have been found in tRNAs, the majority occurring at specific nucleotides in the anticodon stem and loop region.
Modifications fine-tune structural stability, binding affinities, recognition elements and cellular localization signals. They play important roles in proofreading, preventing frameshifting and facilitating elongation factor recruitment during translation.
How are tRNAs transported into the cytoplasm?
Nascent tRNAs are exported from the nucleus to the cytoplasm for use in translation. Export is facilitated by specific carriers and transporters. In eukaryotes, many tRNAs contain a Transport Element that allows recognition by Exportin-t and movement through nuclear pore complexes. Specialized transport factors bind along with Ran-GTP to deliver tRNAs into the cytoplasm where they join the pool bound for aminoacylation. Retrograde nuclear import of aberrant tRNAs provides a quality control checkpoint.
What is the life cycle of tRNAs?
After aminoacylation in the cytoplasm, tRNAs temporarily associate with elongation factors and recruit amino acids to the ribosome for protein synthesis. Each round of translation is estimated to involve over 200 reactive cycles by a single tRNA molecule.
As tRNAs are continually regenerated, they must maintain precise structures and resist degradation. Used tRNAs are disassembled from the ribosome and undergo repair, recycling or degradation depending on the extent of damage. Quality control ensures only intact and fully functional tRNAs support new rounds of translation.
Why is understanding tRNA important for scientific research and human health?
Understanding the complete workings of tRNA is crucial for advancing our knowledge of fundamental biological processes and developing potential therapeutic interventions. By unraveling the mechanisms by which tRNA molecules participate in the translation, researchers can gain insights into the underlying causes of genetic disorders, cancer, and other diseases associated with errors in protein synthesis.
Moreover, the ability to engineer or modify tRNA molecules holds promise for various applications, such as producing biotherapeutic proteins with non-standard amino acids or developing new antimicrobial agents that target the translation machinery of pathogenic organisms.
Summary
In conclusion, while compact in size, tRNAs play a multi-faceted role in gene expression. Their transcription, processing, modification and intracellular distribution involve an array of enzymes and transporters working in exquisite coordination. Continuous regeneration is vital to sustain the high demands of protein synthesis. Tight regulation at each step safeguards the integrity and fidelity of the genetic code. A balanced tRNA pool forms the basis for accurate and efficient protein production in all living cells.